Project as an easy way to manage a study
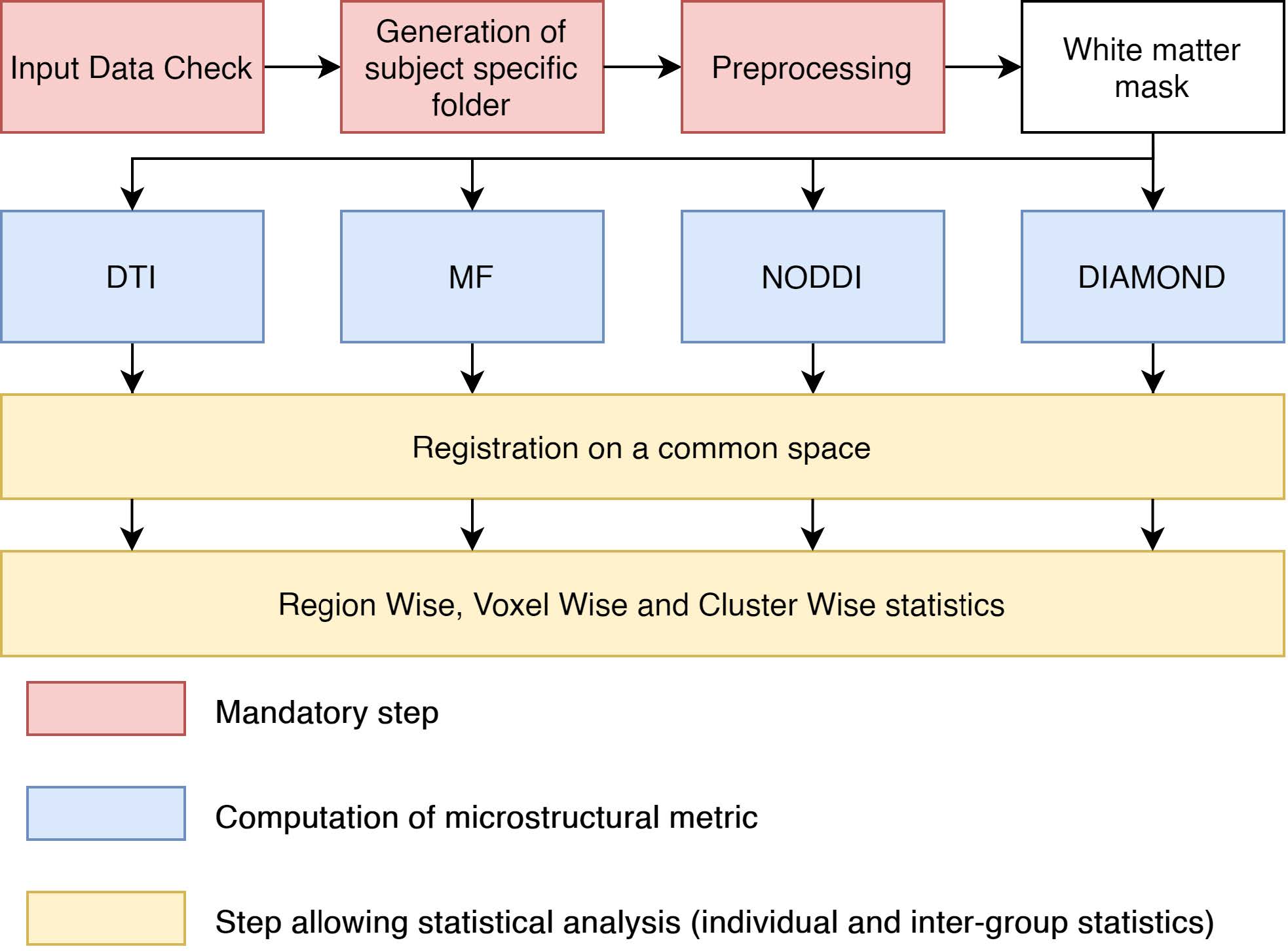
Main available processing steps
Using a rich set of functions, processed data are firstly audited to ensure that the image dimension match the dimension of the b-value and b-vector files along the acquisitions parameters and index files. If successful, a dedicated storage folder for each subject is generated. Afterward, preprocessing can be applied to correct and enhanced the raw image. This includes, brain extraction, reslicing, Principal Component Analysis (PCA) based denoising using the Marchenko-Pastur distribution, suppression in image space of Gibbs ringing artifacts, estimation and correction of the susceptibility induced and Eddy current induced distortions, modeling and correction of subject movements and finally, bias field correction. After the preprocessing, a white matter mask registered on diffusion data can be obtained using a T1 image or computed directly from diffusion data by segmenting an Anisotropic Power (AP) map.
As seen in the figure below, four distinct algorithms for the estimation of the microstructure are available. Subsequently, the pipeline adds the possibility to register the computed metrics in a common space given by either one of the study subject or by an atlas. Finally, from there, ElikoPy can output statistical results for population studies by aligning metrics from multiple subjects into a common space and performing region wise, voxel wise and cluster wise statistics.
Folder structure
The Elikopy toolbox follows a specific folder structure that prevents ambiguity and data losses when dealing with a large amount of subjects. The figure below illustrates the folder tree in ElikoPy.
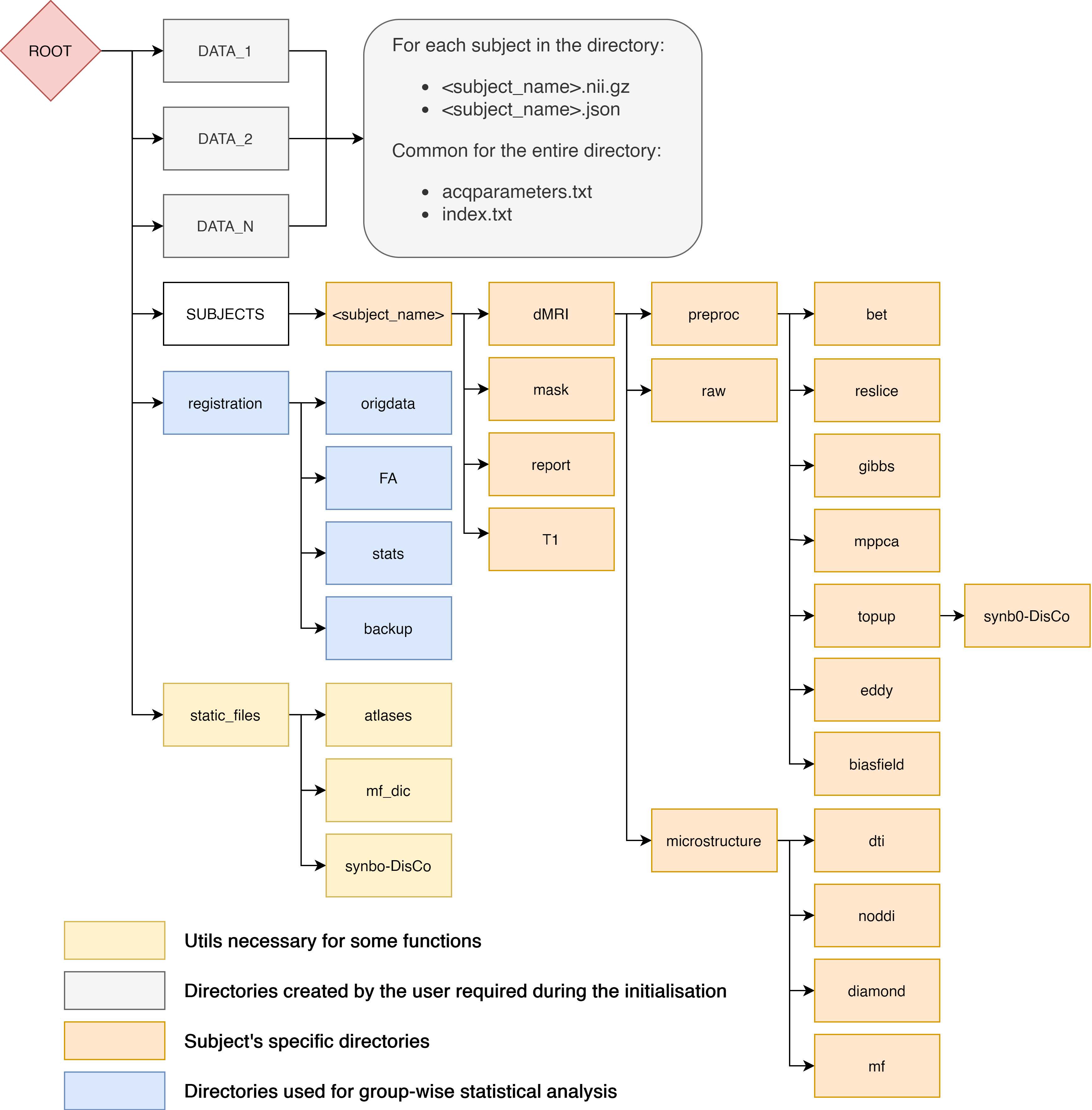
The first type of folder present at the root of the Elikopy project are data folders. These folders contain all raw subject files belonging to the same class along with the associated acquisition parameters and index files. Classes are used to separate subjects that have different acquisition parameters or subjects that need to be separated from others groups of subjects. The pipeline does not have a limitation on the number of classes.
The subjects folder contains a subfolder for each valid subject presents in data folders. Along these subfolders, three json files are present. The subj_error.json file contains the list of subjects with invalid raw data, The subj_list.json file contains the list of valid subjects and the subj_type.json is a dictionary that maps each subjects subfolder to its data class.
Each subdirectory of the subjects folder contains the output of every preprocessing and processing function executed on the patient associated with the subdirectory. The output consists of NIfTI files, log files and some other files related to the specific functions.
The registration folder contains diffusion metrics registered to a common space, group wise statistics and voxel wise statistics for each registered metric.
Finally, the static_files folder contains files mandatory for some processing steps of the library such as MF dictionary and Synb0-DisCo atlases.