Typical usage for processing a study
On this page is presented a basic usage of the ElikoPy library. More information on all these functions are available in the detailed guide.
Header and initialisation
The first step to enable ElikoPy is to import it and initialise the ElikoPy object “study “ specific to the current study. The only required argument for the constructor is the path to the root directory of the project.
1import elikopy
2import elikopy.utils
3
4f_path="/PROJECTS/"
5dic_path="/PROJECTS/static_files/mf_dic/fixed_rad_dist.mat"
6
7study = elikopy.core.Elikopy(f_path)
8study.patient_list()
The root directory must have the following structure during the initialisation
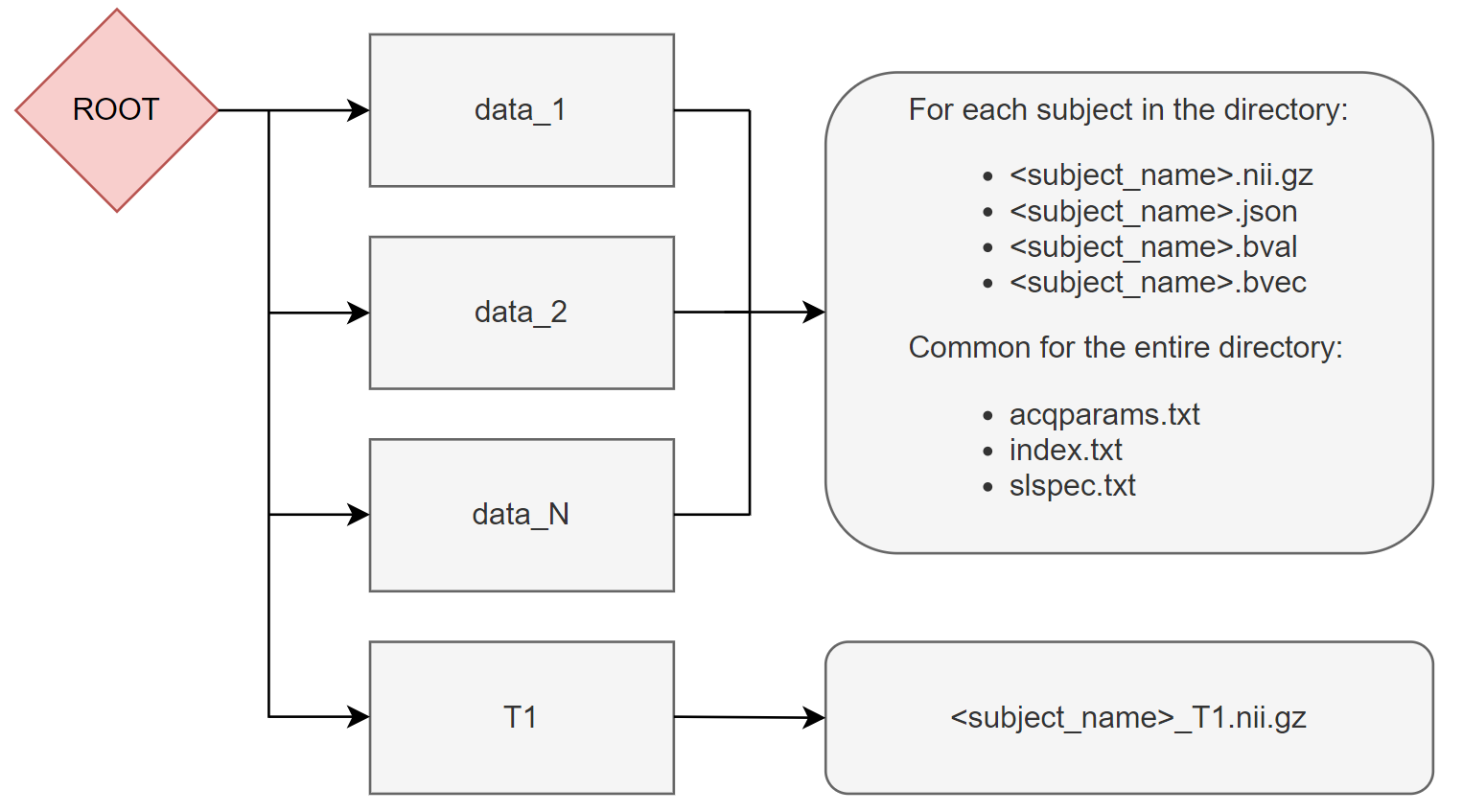
The T1 structural images as well as the acqparams, index and slspec files are optional. However, if they are not available, some processing steps might be not available (this is usually specified by a note in the documentation). These files can be generated as explained in the following links:
acqparams.txt and index.txt : Eddy FSL acqp
slspec.txt : Eddy FSL slspec
Note
If you have volumes that are acquired with reverse phase encoding directions in separate NIFTI files, instead of merging the NIFTIs, it is possible to use the ‘reverseEncoding’ argument of patient_list.
study.patient_list(reverseEncoding=True)
In this case a reverse_encoding directory containing the data with reversed phase encoding direction can be added in the data_N folders and the merging operations will be managed automatically.
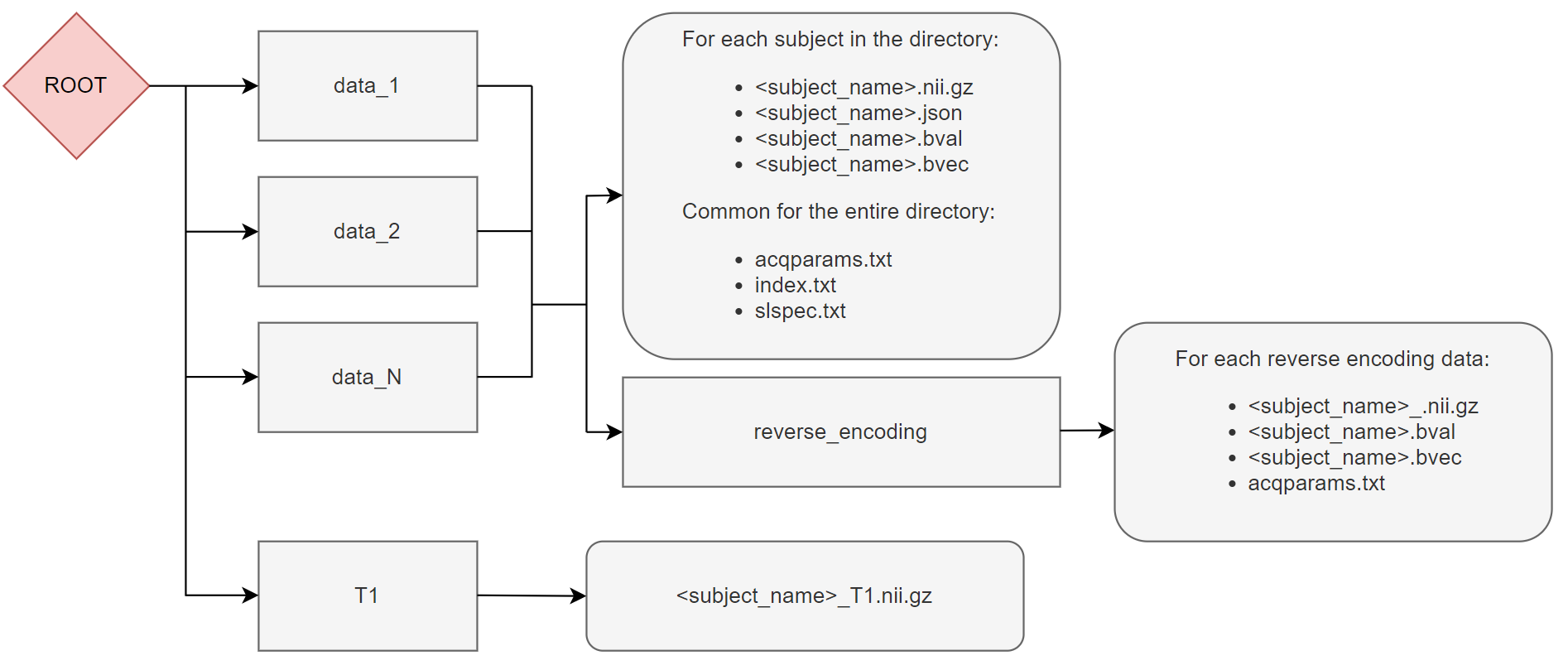
Note
The BIDS format is also supported as input using the bids_path argument of patient_list
study.patient_list(bids_path="path_to_BIDS")
Preprocessing
The following code block shows how to preproccess the dMRI data. By default only the brain extraction is enabled in the preprocessing but we recommend you to enable more preprocessing steps as described in the detailed guide (see Preprocessing of diffusion images).
8study.preproc()
whitematter mask
The following code block computes a white matter mask for each subject from its T1 structural image (if available). If the T1 is not available, the mask is computed using the anisotropic power map generated from the diffusion data.
9study.white_mask()
Microstructural metrics computation
The following code block computes microstructural metrics from the four microstructural models available in ElikoPy.
10study.dti()
11study.noddi()
12study.diamond()
13study.fingerprinting()
Statistical Analysis
In the following code block, fractional anisotropy (FA) from DTI, along with other additional metrics, are registered into a common space. The registration is computed using the FA and the mathematical transformation is then applied to the other metrics.
Afterward, the randomise_all function performs group wise statistic for the defined metrics along extraction of individual region wise values for each subject into csv files.
14grp1=[1]
15grp2=[2]
16
17
18
19study.regall_FA(grp1=grp1,grp2=grp2)
20
21additional_metrics={'_noddi_odi':'noddi','_mf_fvf_tot':'mf','_diamond_kappa':'diamond'}
22study.regall(grp1=grp1,grp2=grp2, metrics_dic=additional_metrics)
23
24metrics={'dti':'FA','_noddi_odi':'noddi','_mf_fvf_tot':'mf','_diamond_kappa':'diamond'}
25study.randomise_all(metrics_dic=metrics)
Data Exportation
The export function is used to “revert” the folder structure, instead of using a subject specific folder tree, data are exported into a metric specific folder tree. In this example, only metrics computed from the dti model are exported.
22study.export(raw=False, preprocessing=False, dti=True,
23 noddi=False, diamond=False, mf=False, wm_mask=False, report=True)
Note
If you wish to learn more about the library and its validation, we recommend you to read the detailed guide and play around with the library.
Other parameters commonly available
The ElikoPy library has been made compatible with the slurm scheduler commonly present on HPC clusters. The use of the slurm scheduler can be controlled with the slurm parameters.
Associated options are:
slurm – Whether to use the Slurm Workload Manager or not (for computer clusters). default=value_during_init
slurm_email – If not None, Topup will use additionnal parameters based on the supplied config file located at <topupConfig>. default=None Email adress to send notification if a task fails. default=None
slurm_timeout - Replace the default slurm timeout used in the ElikoPy function by a custom timeout.
slurm_mem - Replace the default amount of ram allocated to the slurm task by a custom amount of ram.
cpus – Replace the default number of slurm cpus by a custom number of cpus.
The slurm option and slurm_email option can be globally define during the initialisation of the study object.
When processing a study, the processing for some subjects could fail for various reasons. The ElikoPy library provides two parameters destined to limit the amount of processing necessary to recover from these failures.
patient_list_m – Define a subset of subjects to process instead of all the available subjects. example : [‘patientID1’,’patientID2’,’patientID3’]. default=None
starting_state – Manually set which step of the function to start from. default=None